I. Brief description
The isolation and purification of modern plasmid DNA is representative from E. coli. In view of the important position of E. coli in molecular biology research, the isolation and purification of quality control DNA from E. coli has become super An important topic in centrifugal technology. The rapid separation and purification of plasmid DNA places higher demands on ultracentrifugation equipment (ultracentrifuges, rotors and ancillary equipment).
E.coli is a typical prokaryotic organism, because prokaryotic cells lack the inner membrane system of their nuclear cells that can separate multiple functional components into specialized and locally independent regions. There are no organelles (nucleus, endoplasmic reticulum, Golgi apparatus, wire pullers, lysosomes, etc.) contained in their nuclear cells. Electron microscopy shows that E. coli has two distinguishable internal regions, the cytoplasm and nucleoplasm, surrounded by a thin layer of cytoplasmic membrane and a very thick cell wall, with some free flagella attached to the outside of the cell wall. . The plasmid DNA is located in the nuclear region and exists in the form of filaments, which in many cases are polycondensates in which some fragments of extremely long circular DNA are folded.
For the microstructure of E. coli, the pretreatment sequence before ultracentrifugation purification of plasmid DNA is:
E.coli→Using lysozyme to remove cell walls→Using surfactants such as SDS, Triton X-100, etc. to break the cell membrane → Most of DNA, RNA and protein are precipitated with sodium acetate (more than 90%).
The precipitate can be deproteinized by deproteinization on the molecular sieve after adding TE buffer (10m-MTris-HCL 1mM EDTA, pH 8.0). The protein can also be removed by ultracentrifugation, de-RNA, de-graded DNA or DNA fragments. This paper will review the new developments of the latter method in recent years.
Second, the latest development of plasmid DNA ultracentrifugation separation
1. Traditional separation method: A few years ago, due to the limitation of equipment conditions, the separation of plasmid DNA was generally performed by CsCl equilibrium equal density centrifugation and self-forming gradient. Take 10~12ml single-tube capacity as an example, separate it with a flat-turning head, 36.000rpm×60 hours, and use a corner rotor to separate 45,OOrpm×36 hours. The former includes acceleration and deceleration to share the life of 130 million rpm. The latter also needs to use 100 million rpm drive life, which is the total life of the ultracentrifuge at that time is 10-20 billion rpm, no doubt the cost of each experiment is too high, plus CsCl usage, price and other factors, Making such separation and purification work becomes a very expensive experiment.
2. New progress:
(1) Centrifugal separation of overspeed vertical tube rotor (made of alloy or carbon fiber): After the vertical tube rotor was introduced in 1975, the vertical tube rotor developed by major centrifuge manufacturers in recent years has a single tube capacity of 0.2. From ml to 4Oml, the maximum speed is from 50,000 rpm to 120,000 rpm, and the RCFmax can reach 700,000×g. The new models and rotors developed in the 1990s have made it possible to make the vertical separation of plasmid DNA into experiments.
The point of separation and purification of plasmid DNA using a vertical tube rotor is:
· Due to the shortest settling distance, the shortest centrifugation time, high efficiency, low consumption:
·The longitudinal section area of ​​the centrifuge tube is far and large, and the transverse section area is large. The capacity of the pure Xiangpin zone is large when it is centrifugally settled. The separation purity is high under the condition that there is no sedimentation or the precipitation is tight and dense, and the sample loading amount is also large; Hydrostatic pressure, the biological particles will not be damaged due to excessive static pressure. The hydrostatic pressure of the sample particles in the centrifuge tube is:

Where: ω: angular velocity
r: the distance between the sample particle and the center of rotation (in centimeters)
R1: The minimum centrifugal radius of the rotor is rmin (cm), which is the distance between the liquid level and the center of rotation when using gmax centrifugation.
Pa: initial density (minimum density for pre-formed gradients, centrifugation for self-forming gradients, minimum density at the end of centrifugation).
a: gradient slope
In ultracentrifugation, the P value is quite large. It is generally considered that P≤1500kg/cm2, so as not to damage the biological particles, the vertical tube rotor (r-r1) is small, and relatively speaking, P is also quite small (102 orders of magnitude). When turning the head, it is often necessary to check the P value before centrifugation. When it is too large, the speed should be reduced. When the plasmid DNA is isolated and purified by a vertical tube, the RNA precipitate is attached to the wall of the centrifuge tube. When decelerating or gradient direction is switched, the plasmid DNA band is "wiped" from the precipitation zone, and a small amount of RNA is mixed into the DNA to affect the purity.
At very high speed centrifugation (eg, greater than 80,000 rpm), this effect is less due to the tighter attachment of the RNA to the wall.
(2) Centrifugal separation of the near vertical tube rotor: in order to eliminate the contamination of the formed DNA band by the RNA precipitation formed in the wall by the vertical tube rotor for plasmid DNA centrifugation, and also to improve the general bevel type rotor ( Inclination angle 25·~35·) Due to the long settlement distance and the long separation time, a variety of near vertical tube rotors have been developed in recent years (ie, Near VerticalTube Rot, referred to as NVT rotor or Neo Angle Rotor). Small false angle rotor, referred to as NT). The angle between the central axis of the longitudinal section of the centrifuge tube and the drive axis of the centrifuge is between 7.5·~10·, the speed is from 65,000 rpm to 120,000 rpm, and the RCFmax is up to 646,000×g. The single tube capacity ranges from 2ml to 13.5ml. The development of NVT (or NT) rotors is mainly designed for the isolation of plasmid DNA. Of course, it is also suitable for the separation and purification of mitochondrial DNA, chromosomal DNA, RNA and serum lipoprotein.
The characteristics of the purified plasmid DNA using the NVT (NT) rotor are:
The time required for centrifugation is slightly longer than the vertical tube rotor (30% to 40% increase), but shorter than the general angled rotor (60% to 70% of the separation time of the same angled rotor).
Although the outer wall of the RNA plate centrifuge tube has a small sliding force due to the small angle of inclination, after the addition of a surfactant (such as 0.1% to 0.001% Triton X-100), the RNA precipitation can slide to the bottom of the centrifuge tube faster (ie, In the self-forming gradient, the RNA has reached the bottom), and the purity of the plasmid DNA is not affected during the gradient conversion;
The hydrostatic pressure is small, and the biological particles are not damaged when running at a high speed;
The longitudinal section of the centrifuge tube is larger than the general angle type and the flat head, and the separation purity is high, and the sample is added in a large amount.
(3) Discontinuous step gradient separation: The traditional method for separation and purification of plasmid DNA is to use a full-tube CsCl self-forming gradient equilibration equal density centrifugation method. The density of the whole tube CsCl is uniform at the beginning of centrifugation, and the sample is evenly distributed.
The centrifugation time of the CsCl self-forming gradient can be calculated by the following formula:

Where N: actual speed (rpm)
P: Floating density of sample (such as plasmid DNA) in CsCl
r: the distance (cm) from the center of rotation to the center of the pure sample strip. As a preliminary settlement, the plasmid DNA is centrifuged, and the r position can be set in the middle of the centrifuge tube, and the r average is 10%. (rmax-rmin).
F: Constant coefficient depending on the gradient material and the initial density of the solution <Table 1>.
S20.w: The sedimentation coefficient of the sample (plasmid DNA) in water at 20 ° C can be determined by reference to the relevant literature.
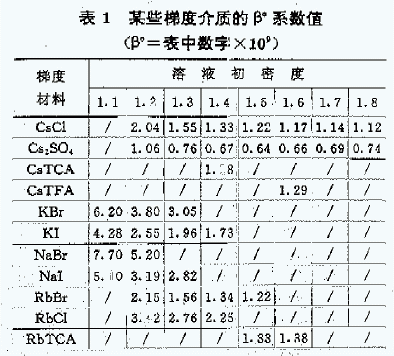
[Note] TCA in the table is trichloroacetic acid, TFA is trifluoroacetic acid
The centrifugation time calculated by the above formula is very close to the actual centrifugation result. It can be seen from the formula that the T inverse ratio (N4, r2), obviously increasing (N4, r2) under the premise that CsCl does not crystallize will reduce the centrifugation time, and the effect of increasing the rotational speed is particularly obvious. However, for some lower-speed rotors (such as below 65,000 rpm), the time required for CsCl self-forming gradient is too long (10 hours), so the formation time of the pure sample of plasmid DNA is also longer.
As an improvement to the initial isocratic conditions of the whole tube, in recent years, the equilibrium equal density centrifugation of the stepped discontinuous CsCl gradient has been developed, that is, the initial gradient of the centrifuge tube is divided into two parts:
Lower part: high density area (Ï=1.80-1.81g/cm3), accounting for 1/3 of the total capacity.
Upper part: low density zone (Ï=1.46-1.48g/cm3), accounting for 2/3 of the total capacity. Experiments have shown that the second-order discontinuous CsCl gradient can be much shorter than the time required for full-tube iso-density centrifugation (with 12ml vertical tube). For example, the transcript, plasmid DNA, CsCl gradient 55,000 rpm, 20 ° C; discontinuous two-step centrifugation for 5.5 hours, whole tube equal density centrifugation for 8 hours).
Second-order discontinuous gradient centrifugation, the sample is applied to the high-density area at the bottom of the centrifuge tube, which is very large regardless of the type of rotor; and there is only the sample floating up without the low-density area (low-centrifugal acceleration zone) sample. The sedimentation; the initial density difference is large in the near-DNA floating density region, the self-forming gradient is shorter, and the centrifugation time is shortened due to the above reasons. (4) Super high-speed multi-stage (speed) separation: Obviously, increasing the speed according to the T formula will accelerate the formation of the CsCl gradient, and the centrifugal time will be shortened accordingly. However, at high temperatures at a certain temperature, long-time centrifugal separation is likely to cause local crystallization for higher density CsCl, while local precipitate precipitation will not only affect the gradient, but also due to the precipitation of solid CsCl due to damage to the centrifuge tube. The wall causes a centrifugal failure or an accident. For this reason, the plasmid DNA centrifugation experiment is characterized by the programmable operation of the novel overspeed machine. It can be gradually moved from very high speed to slightly lower speed. Each experiment is divided into many speed blocks, which improves the efficiency and ensures the success and safety of the experiment.
Of course, this type of experiment is going through several explorations to form a common centrifugation program. Both Beckman and Hitachi Koki Co., Ltd. have done a lot of experiments on their advanced overspeed machines, and recommended reliable and efficient separation to users. Psychological order [2, 3].
Third, typical plasmid DXA ultracentrifugation separation example
1. Traditional separation example:
Example (1) The angle of the single tube is 12ml, the angle of rotation is below 50,000 rpm, the 12PA sealed tube, and the new super speed machine of each factory.
Sample + TE solution (formulated as pH 8.0), EB addition amount 0.2 mg / ml, adding CsCl to form a whole tube Ï = 1.57g / ml 45,000 rpm × 36 hours, 20 ° C, slow deceleration or 55,000 rprn × 16 hours + 40,000 rpm × 1 hour, 20 degrees slow deceleration.
Separation results: a pure plasmid DNA band was formed slightly in the middle of the tube, a DNA fragment band appeared slightly in the middle, and RNA was precipitated near the bottom, and the floating material was protein.
Features: The separation result is better, but the pure sample strip is wider, the separation time is very long, and the life of the drive unit is nearly 100 million rpm.
Example (2) Iron tube with a single tube capacity of 5 ml and a maximum speed of 65,000 rpm, 5PA tube, sample configuration (1)
Features: Same example (5) (use the life of the drive unit of 0.2 billion rpm).
36,000 rpm x 55 hours, 20 ° C, or 32,000 rpm x 70 hours, 20 °C.
· Separation results and characteristics: The separation result is ideal, the plasmid DNA band is narrow, and the RNA is deposited on the bottom of the centrifuge tube. However, the centrifugation time is too long and the cost is high (use 110 to 130 million rpm drive life).
2. Vertical tube rotor centrifugal separation:
Example (3) Japan Hitachi cp100α host, P100VT rotor (700,000 × g, 8 × 5ml), 5PA sealed tube, sample and gradient configuration same example (1)
100,000 rpm × 1 hour 50 minutes, 20 ° C, slow deceleration (7 steps).
·Characteristics: The results of centrifugation are similar to those of the case (1). Due to the extremely high rotational speed, the precipitation of the wall RNA is very tight, and there is little DNA contamination during the deceleration. The centrifugation time is short, efficient, and low-cost (using a life of 12 million rpm).
Example (4) Titanium vertical tube rotor with a maximum speed of 50,000 rpm, capacity 8 x 40 ml, 40PA sealed tube, sample configuration as in (1) 50,000 rpm x 24 hours, 20 ° C, slow deceleration.
Features: Large volume separation, RNA precipitation is slightly contaminated with DNA bands.
3. Near vertical tube rotor separation
Example (5) Beckman1 NVT90 rotor XL-90 mainframe, 8 x 5 ml, 5PA sealed tube, rotor top speed 90,000 rpm, 646,000 x g.
Sample + TE solution (formulated as pH 8.0), E·B was added in an amount of 0.2 mg/ml, Triton X-100 was added in an amount of 0.01%, and Cscl was added to prepare p = 1.55 g/ml. 78,000 rpm × 4 hours, 20 ° C, slow deceleration.
Features: Due to the action of Triton X-100, RNA precipitation accelerates to the bottom of the centrifuge tube, and there is no pollution to the DNA during the deceleration of the rotor. The separation result is ideal (use 20 million rpm drive life).
Example (6) Japan Hitachi CS120EX or US Beckman TLX micro overspeed machine, NVT (NT) rotor with a maximum speed of 120,000 rpm, 8 × 2 ml, 2PA sealed tube, sample and gradient liquid configuration basically the same example (5), but CsCL Formulated as p = 1.55 g / ml, 120,000 rpm × 3 hours, 20 ° C, slow deceleration.
Features: the same example (5) (use 20 million rpm drive life)
4. Discontinuous step gradient separation:
Example (7) Japan Hitachi SRP83VT rotor, 80,000 rprm, 549,000g, 8×5, 5PA sealed tube (Hitachi CPα, β series machine or SC summer series machine), gradient liquid configuration:
First configure TE liquid + Cscl, Ï = 1.47g / ml a total of 3.5ml, first injected into the 5PA sealed tube.
Then configure TE solution + sample + CsCl to form Ï = 1.81g / ml, EB 0.2mg / ml, a total of 1.5ml with a syringe into the bottom of the tube slowly injected to make the original p = 1.47 liquid floating.
83,000 rpm × 1 hour, 20 ° C, slow acceleration, slow deceleration, the same result (3).
Features: Due to the use of second-order discontinuous gradients, the CsCl self-forming gradient time is shortened. Ultra-high speed, RNA precipitation is very tight, DNA contamination is very small, efficient, low cost (only use 0.05 million rpm drive life). The disadvantage is that the amount of sample added is slightly smaller.
Example (8) Japan Hitachi RP55VF: rotor, 55,000 rpm, 293,000 × g, 12 × 5 ml, sample and gradient liquid configuration same (7) 55,000 rpm × 4.5 hours, 20 degrees, slow addition, deceleration. The result is similar to the case (7).
5. Super high speed multi-stage (speed) separation:
Example (9): Beckman XL-90 overspeed machine, NVT-90 rotor 90,000 rpm, 645,000 x g, 8 x 5 ml, 5.1 PA sealed tube, sample and gradient solution configuration (5).
Multistage separation: 90,000 rpm x 1.5 hours + 87,000 rpm x 0.25 hours + 83,000 rpm x 0.25 hours + 81,000 rpm x 0.50 hours + 80,000 rpm x 0.50 hours, 20 ° C, slow deceleration.
(The above experiment can also be done with the P100VT rotor on the Japanese Hitachi CP100α or 90α mainframe, the results are the same).
Features: The result is the same as in the example (5), but the time is reduced to 3 hours (16 million rpm)
Example (10) Hitachi micro-speed CS-120EX or CS-100EX micro-speeding machine, S100AT5 angle rotor, 100,000 rpm, 550,000 × g, 8 × 5ml, sample and gradient liquid configuration is basically the same as example (3), but the sample +TE solution is formulated into Ï= 1.55g/ml
100,000 rpm × 4.5 hours + 98,000 rpm × 15 minutes + 96.000 rpm × 30 minutes + 94,000 rpm × 30 minutes + 90,000 rpm × 25 minutes + 85.000 rpm × 30 minutes
(7 hours total), 20 ° C, slow deceleration.
Features: Using angled rotor, micro-speeding machine, under the condition of small centrifugal force (compared with large overspeed machine), the centrifugation time is shorter, RNA sinks to the bottom of the tube, and the separation result is ideal.
Fourth, the consideration of plasmid DNA ultracentrifugation separation
1. Prevent pollution and prevent dehydrogenase contamination: Dehydrogenase can degrade or denature DNA, and it is ubiquitous (on the skin, the surface of the device that has not been cleaned and not disinfected...), so in the plasmid DNA separation experiment Pay attention to this in every step. The effective method is disinfection of the appliance (centrifuge tube, cap, syringe, pipette, container, etc.) (steam disinfection or 0.1% diethylpyrocarbonate disinfection). Although we can add DFP (diisopropyl acid fluoride) or PMSF (benzyl fluorophosphate) to inhibit the degradation of dehydrogenase, the main means is cleaning and disinfection.
To prevent ribosome contamination on the skin, surgical gloves should be worn during operation.
2. Prevent the heavy metal ions and DNA present in the heavy metal salt from binding into a complex: CsCl is an ionization medium. CsCl should be passed through the column to eliminate Cs ions before centrifugation, and the vessel contacting the sample should also be washed and rinsed with deionized water.
3. Sample Loading The amount of sample added is related to the sample concentration. In order to improve the resolution, the amount of sample added should be controlled. The appropriate amount of addition should be used as a reference for previous experiments. When self-testing, the sample volume per ml of gradient solution is about 10μg~20μg.
4. For the self-forming gradient balance and other density separation of angle, vertical and near vertical rotors, it can be accelerated by 0~100Orpm and slow decelerated by 1,000rprn~0. The step gradient requires slow acceleration and slow deceleration. Provides a good example for users to choose when adding and decelerating rates.
5. The temperature of the rotor should be basically the same as the temperature at the start of centrifugation (5 °C), the pre-cooling temperature of the rotor is too low (below 10 °C). At very high speed, when it is centrifuged for a short time, it may not be warmed up. The precipitation of CsCl occurs, causing the experiment to fail or an accident.
6. Appropriate sampling method: It is simple to carry out ultracentrifugation of plasmid DNA, and the sampling method and operation level after centrifugation will determine the recovery rate and sample purity. First, the sampling environment should be similar to the centrifugal working temperature (±5 °C); there should be no vibration source on the test bench when sampling; transverse puncture, bottom void, centrifuge tube cutting or gradiometer collection according to laboratory conditions. But no matter which method department must be careful.
7. Appropriate pH value, nucleic acid products should be kept in TE solution of pH 8.0 throughout the experiment. If the pH is too high or too low, DNA will be denatured.
8. Centrifuge tube and dosing amount: Because the ultracentrifugation of the modern quality DNA uses very high rotation speed, the material of the centrifuge tube is also very high. Generally, the centrifuge tube for such experiment is used only once. It is best to use a sealed tube, and a section to select a PA tube is preferred. The storage period of the centrifuge tube should not exceed 2~3 years, and the sample should be filled up with a sealed tube or a thin-walled tube with a cover. Use a flat-turn rotor head 2 to 3 mm from the nozzle to prevent meniscus spillage or nozzle rollover.
â… .DESCRIPTION
Aluminum Bitumen Butyl Tape is a cold applied self adhesive aluminum backed flashing in tape form.
â…¡.USES
Window and door openings (headers, sills,jambs, thresholds, nailing flanges)
Deck-to-wall intersections
Corner boards
Wall-to-wall tie-ins
Foundation sill plates
Sheathing panel seams
Under stucco finishes
Masonry walls
Roof detail areas
Gutters
Mobile home repair
Other building joints.
Exposed pipelines protection.
â…¢. COMPOSITION
High melting point polymer bitumen adhesive laminated to a 30-70 micron aluminium. The adhesive surface is protected by a release coated plastics film which is discarded during use.
â…£.Features
Easy application-Installation fast and easy-simply remove the release film and press onto the substrate.
Superior adhesion capabilities-Creates a strong bond to the substrate for long-lasting waterproofing
Protection.
Excellent sealing performance-The specially formulated rubberized asphalt adhesive seals around fasteners, allowing no water to penetrate and get to the substrate.
Highly conformable and flexible-Can accommodate settlement and shrinkage movement.
Long-lasting waterproofing protection-Both the aluminum surfaced polyethylene film and pure alu surface with the specially-formulated rubberized asphalt components create a water and moisture barrier that does not degrade from the effects of the environment.
â…¤.Technical Datas:
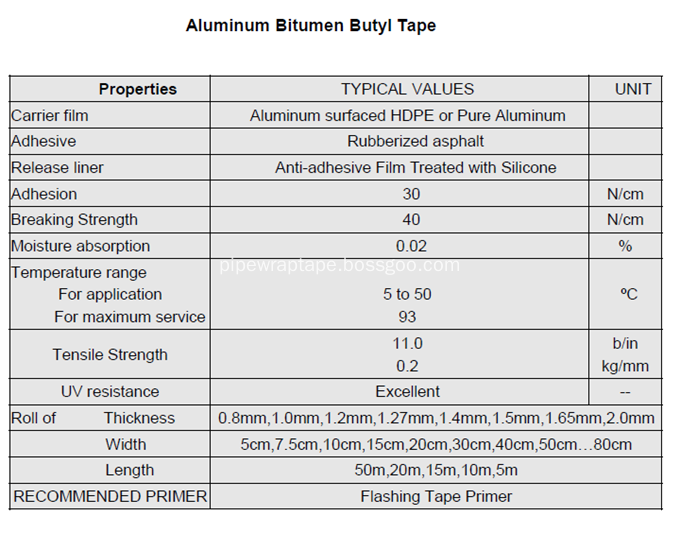

Waterproofing Wrapping Tape, Waterproof Aluminium Bitumen Flashing Tape, Alu Flashing Tape
Jining Xunda Pipe Coating Materials Co.,Ltd , https://www.pipe-wrap.com